Using Data Files
[Genhelp | Program Manual | User's Guide | Data Files | Databases | Release Notes ]
Table of Contents
Overview
What are data files?
Default vs. local data files
Using local data files
Creating or modifying data files
Using a special kind of data file: A scoring matrix
Using scoring matrices
Converting scoring matrices to a different format
Converting pre-version 9 scoring matrices to the new format
Converting BLAST-format scoring matrices to GCG format
Overview
This section explains how Accelrys GCG (GCG) programs work with data files. Data files contain non sequence information which some programs need to perform their analyses.
You are not required to create or specify data files to successfully use GCG programs. All programs that require a data file have a default file they use, so as a new user you needn't worry about the information in this section.
This section is for intermediate to advanced users who understand how programs access data files and who want to modify them or create their own files to customize their analyses. You'll learn how to
- Specify and use local data files with GCG programs.
- Access default and alternative public data files and modify them for your personal use.
- Work with a special kind of data file, a scoring matrix.
Note: Plus (+) programs in the GCG are enhanced versions of the corresponding non-plus programs of releases prior to 11.0; example: BLAST+. In addition, there are five plus programs that are new to this version. For example: ClastalW+.
Plus programs prompt you for information: they usually require a sequence for input, a name for an output file, the beginning and ending positions of the portion of sequence you want to use, and other pieces of information particular to individual programs.
Several of the enhanced “plus” programs use data files, but none of the plus programs are able to use local data files or logical names.
What are data files?
By now you've learned the basics of how
to use GCG programs to analyze the nucleic acid or protein sequences that
are stored in the sequence databases or in your own personal sequence files.
Additionally, many programs require nonsequence information, or data files,
which they use to analyze the sequences. For example, one of the nucleic acid
mapping programs, Map+ requires two data files:
enzyme.dat, which contains restriction enzyme names and their corresponding
recognition sites; and translate.txt, which associates codons with their
corresponding amino acids.
Default vs. Local data files
Default data files
All programs that require a data file
have a default file they use, so as a new user, you need not worry about
supplying one. These default files are public--that is, they are available to
everyone who uses the package. Default data files are located in the public directory
with the logical name SHARE. When you run a program that requires a data file,
it automatically finds the appropriate default file in this directory; this
means you don't have to specify the directory and filename.
GCG also includes alternative data
files you can use with a program instead of the default file. There may be
times when you want to use an alternative data file rather than the default
one. For example, if you're using the CodonPreference
program to analyze a Drosophila sequence, you may want to use the alternative
codon frequency table drosophila_high.cod rather than the default table,
eco_high.cod, which is more appropriate for bacterial sequences. These
alternative data files are located in the directory with the logical name
SHARE. You can then select the appropriate data file from the relevant
directories (codon, energy, matrix, etc)
Local data files
You also can create your own data files,
or you can copy a default or alternative public data file to your local
directory and modify it to suit your needs. These files are known as local
data files. For instance, let's say you're working with the Map program and you create a data file of enzymes specific
to your research. This personal data file, then, would be available only to
you. When you have a local data file a program can use, the program tells you
so with a message similar to *** I
read your "data" file
*** to remind you that
you have a data file in your directory that the program is using instead of the
default file.
How do I know what data file a program uses?
You can find what default data file a
program uses in a number of places:
Program Manual. Check the "Local Data Files"
topic toward the end of each program entry in the Program
Manual. This topic provides you with a summary of how data files work and
briefly describes each default data file used by the program. The "Command
Line Summary" topic of each program in the Program Manual also lists any
default data files the program uses.
In addition, you can find default and
alternative data files listed and described in Appendix
VII of the Program Manual.
Online documentation. Display the default data file(s) a
program uses by running the program with the -CHEck
parameter. The program displays the command-line parameters, including the
default data files and parameters you would use to specify alternate data
files.
Data file information is also available online
in GenHelp. You can find default data files listed in each program within the
subtopic "Local Data Files."
Ways to Specify Data Files
Local versions of data files are always
optional; you are never required to supply one because there is always a default.
However, if you choose to provide a local data file or an alternative public
data file, you can do so in a number of ways. GCG programs have a hierarchy
of locations they check for data files.
Most programs check for data files in the
order described below. (Scoring matrices use a different search order,
described in more detail in "Using
a Special Kind of Data File: A Scoring Matrix" later in this section.)
- On the command line. Programs check on the command line first to see if you specified a data file using a parameter, for example, -DATa=filename or -TRANSlate=filename. If the data file is not in your working directory, you must specify the directory path. (See the "Local Data Files" subtopic of each program in the Program Manual or online help for the specific parameter you will need.)
- In your working directory. If you did not specify a data file on the command line, programs will check in your working directory for a file with the same name as the default data file. For example, the default data file for the PeptideMap program is proenzyme.dat. If you had a file in your current directory with the name proenzyme.dat, the PeptideMap program would automatically use it instead of the default file.
- In the directory
with the logical name MyData.
If the program did not find the appropriate file in your working
directory, it then will check for a directory with the logical name
“MyData”.
If you frequently use alternative data files or have modified or created your own data files, it is a good idea to set up a directory with the logical name MyData and place all of your local data files in that directory.
- In GCG default
directory. The last place a program looks for a data file is in SHARE, where it
always finds the default data file.
The “$GCGROOT/share/” data
directory is a special feature within the package. Because programs
automatically search for the logical name SHARE, you need not worry about what
directory you are in when you run a program that uses a data file. The program
automatically finds the data directory. For more information about defining
logical names for directories, see "Defining
and Using Logical Names for Directories" in the "Working
with Directories" section of Section 1, Getting
Started.
Types of data files
There are many different types of data
files you can use to customize a program's analysis. For more information about
these data files, see Appendix VII of the
Program Manual.
- PROSITE. Used by the Motifs program, this data file lists the sequence motifs in the PROSITE Dictionary of Protein Sites and Patterns, distributed by the European Molecular Biology Laboratory (EMBL).
- Profiles. ProfileScan uses a table of validated profiles derived from the motifs in PROSITE.
- Codon Frequency. Several GCG programs use a table of codon frequencies to make some inferences about the probability of codons occurring in a nucleotide sequence. The package provides codon frequency tables for Drosophila, Human, Maize, and Yeast as well as E. coli highly expressed genes, which is the default.
- Translation. These tables serve three purposes: 1) to define the relationships between codons and amino acids; 2) to define the relationship between one-letter and three-letter amino acid codes; and 3) to identify potential start codons and stop codons. To specify an alternate translation table on the command-line, use the parameter -TRANSlate=filename.
- Pattern. Several GCG programs use pattern files, which define one or more patterns that a program searches for. You can create your own pattern data file or use one of the following types:
Restriction
Enzymes (REBASE). GCG mapping programs Map, MapSort, and MapPlot read
restriction enzyme names, recognition sites, cut positions, and overhangs from
a restriction enzyme data file.
Proteolytic
Enzymes, and Reagents. GCG peptide mapping programs PeptideMap
and PeptideSort require a data file that lists
peptidases and proteolytic reagents and the residues at which they cleave.
Transcription
Factor Recognition Sites. FindPatterns, Map,
MapSort, and MapPlot can optionally use this data file, which lists the
recognition sequences for eukaryotic sequence-specific transcription factors.
- Scoring Matrices (formerly known as symbol comparison tables). These matrices provide a numeric value for each pair of bases or amino acids compared. For example, a matrix might assign a value of 1 for matching symbols and a value of 0 for mismatching symbols. If you compared amino acids, the matrix might assign a number based on chemical similarity or evolutionary distance. The number might be negative if two residues were very dissimilar. Any symbol comparisons not accounted for receive a value of 0. To specify an alternate scoring matrix for a program, use the parameter -MATrix=filename.
Scoring
matrices follow slightly different rules than other data files. For more
information, see "Using a
Special Kind of Data File: A Scoring Matrix" later in this section.
- Protein Analysis. The programs that analyze proteins (see the functional table of contents in the Program Manual) require tables that contain data for predicting peptide isolation, secondary structure, hydrophobicity, antigenicity, isoelectric point, molecular weight, and extinction coefficients.
- Energy. Used by the MFold and Prime programs, these tables contain stacking and loop destabilizing energies.
Using local data files
Data files are local when they
are located in your directory. Local data files may be files you created, or
they may be public data files you copied to your local directory to modify and
use. When you have a local data file a program can use, the program tells you
so with the message
*** I read your "data" file. ***
This message reminds you that you have a
data file that the program is using instead of the default.
To use a local data file:
Choose one of the following.
- Specify the local data file you want to use on the command line with the appropriate parameter, for example -DATa=filename or -TRANSlate=filename. If the file resides in a directory other than the one you are currently working in, you also must supply the directory path. You can find the appropriate parameters to use with data files by adding -CHEck on the command line when you run the program. For example
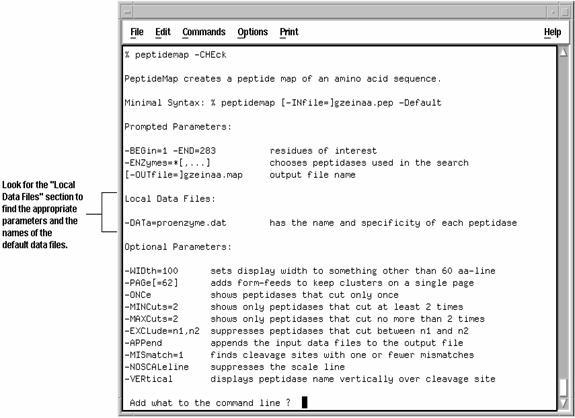
You also
can find the parameter(s) listed in the "Local Data Files" section of
each program in the Program Manual and
Command-Line Summary. In addition, this information is available in the online documentation
GenHelp.
If the file
is in a directory other than your current directory, specify the directory
path: -DATa=/directory/filename, for example -DATa=/project/my_enzyme.dat. If the file is in a directory with a
logical name, specify the logical name followed by a colon and the filename: -DATa=logical_name:filename, for example -DATa=proj:my_enzyme.dat.
- For the program to automatically use a local data file, place the data file in the directory in which you will run the program. You must give the data file the same name as the default data filename. For example, the default data file for the PeptideMap program is proenzyme.dat. If you had a file in your current directory with the name proenzyme.dat, the PeptideMap program would automatically use it instead of the default file.
You can
find the default data filename by adding -CHEck
on the command line when you run the program. You also can find this
information listed in the "Local Data Files" topic of each program in
the Program Manual and in the online documentation GenHelp.
- Create the MyData directory to contain and organize your local data files.
- Create a directory
with the logical name MyData. For more information, see "Defining
and Using Logical Names for Directories" in the "Working
with Directories" section of Section 1, Getting Started. For
example
% cd
% mkdir datadir
% setname MyData ~/datadir
Note: To save your logical names from one
session to the next, add the logical name definitions to your dblogicals.conf
in /HomeDir/ .wp directory). For more information, see "Defining Logical Names" in the
"For Advanced Users" section of Section 1, Getting Started.
- Create or copy the
data file into the MyData directory. For example
% cp data_file datadir
- Verify that the name of the data file you want to use is the same name as the default data filename. You can find the default data filename(s) by adding -CHEck on the command line when you run the program. You also can find the parameter(s) listed in the "Local Data Files" section of each program in the Program Manual and in the online documentation GenHelp.
When you
run a program, it automatically checks if you have a Data directory
(“$GCGROOT/share/”) and uses the data file with the same name as
the default data filename.
Note: When you place a data file in MyData and
rename it to the default data filename, all programs that require a data file
with that name automatically use it each time you run the program. Make sure
this is what you intend before placing files in MyData.
TIP - The Wisconsin Package Version 10.3 (and older versions) included default and alternative data files for you to use. Because data files require a special format, you may find it easier to modify one of these files rather than create a local data file from scratch. In GCG only one path has been provided for all the data files. You can find the default data files in the directory with the logical name SHARE. For more information, see "Creating or Modifying Data Files" in this section.
Creating or modifying data files
GCG includes default and alternative
data files for you to use. However, there may be times when you want to create
a new data file or modify an existing one to customize it to your needs. For
instance, you may want to create your own customized enzyme data file
containing only the restriction enzymes specific to your mapping project.
Because data files have a particular format they must follow, we suggest that
if you want to create a new data file, you should use an existing data file as
a template. You can do this by using the Fetch program
to copy the data file to your directory and then modifying it with a text
editor. Once you copy the file to your directory, it becomes a local data file.
To modify a default or alternative public data file:
- Move to the directory you want to contain the data file.
- Use the Fetch+ or Fetch command to copy the public data file to your current directory. Type % fetch filename, for example % fetch enzyme.dat. A copy of the file appears in your directory.
- Edit the file in the text editor of your choice, for example vi.
Note: All data files require a specific format.
Most data files, such as translation tables, scoring matrices, codon frequency
tables, protein analysis files, and energy tables, require two periods (..)
between the documentary heading and the table itself. In addition, all data
files supplied with GCG have a file type, for example !!CODON_ FREQUENCY 1.0, that appears on the first line of the
file. Do not edit or delete this line. For more information about data file
formats, see Appendix VII of the Program Manual.
- Save the file and exit from the text editor.
To use the
modified data file with a program, see "Using
Local Data Files" in this section.
Using a special kind of data file: A scoring matrix
A scoring matrix is a table of pairwise
relationships between nucleotide symbols or between amino acid symbols. These
tables are used by several programs, including database searching and multiple
sequence alignment programs. In many ways scoring matrices are like other types
of data files used by GCG. However, there are some differences covered in
this section that you will want to note.
Types of scoring matrices
GCG works with two types of scoring
matrices: native GCG matrices and native BLAST
matrices. You can find native GCG scoring matrices in the directory
“$GCGROOT/share/matrix”. If you want to use a native
BLAST-formatted scoring matrix, you can use it directly with a GCG program
without first converting it to GCG format. However, there are reasons you may
want to convert native BLAST matrices to GCG format:
- By default GCG assumes all native BLAST scoring matrices are protein. Because gap creation and extension penalties are calculated differently depending on if the matrix is nucleotide or protein, you may want to convert the native BLAST matrices to ensure they are the correct type. To convert protein BLAST scoring matrices to nucleotide, you can use the Reformat program (see "Converting BLAST-Format Scoring Matrices to GCG Format" in this section for more information).
- If you use native BLAST scoring matrices with GCG, programs determine gap creation and extension penalty values on the fly. However, if you convert a BLAST matrix to GCG format, you can set specific gap creation and extension penalties within the scoring matrix file.
Ways to specify scoring matrices
Using a scoring matrix is similar to how
you use other data files with GCG programs. Each program that uses a scoring
matrix has a file it uses by default, so you are never required to supply one.
However, using scoring matrices differs from using other data files in two
ways. 1) You use a different parameter, -MATrix=filename, to specify an alternate scoring matrix on the command
line. And 2) if you choose to provide an alternate scoring matrix on the
command line, GCG uses a slightly different search order for finding the
file you specify. If you specify the directory where the scoring matrix
resides, the package looks only in that directory. For example, -MATrix=./project/pam250.cmp looks only in the /project subdirectory for the file pam250.cmp. However, if you specify the filename
alone, for example -MATrix=pam250.cmp, the ackage looks for that file in the
directories described below. (In contrast, -DATa=filename looks for the file only in your current directory or in the
directory you specify.)
- In your working directory. Programs will check first in your working directory for the scoring matrix you specified.
- In the directory with
the logical name MyData.
If the program did not find the appropriate file in your working
directory, it then will check for a directory with the logical name
“MyData”.
If you frequently use alternate data files or have modified or created your own data files, it is a good idea to set up a directory with the logical name MyData and place all of your local data files in that directory.
- In GCG default directory ($GCGROOT/share/matrix).
If the program did not find the specified scoring matrix in your working
directory or the directory with the logical name “MyData”, it
then will check for matrix file in the default directory.
Using scoring matrices
To specify an alternative scoring matrix:
Use the parameter -MATrix=filename, where filename
is the name of a scoring matrix residing in 1) your current directory, 2) the
directory with the logical name you have specified in the earlier step
Converting scoring matrices to a different format
There are a couple of reasons why you
might want or need to convert scoring matrices:
- You must convert all local pre-version 9 scoring matrices to the format implemented in version 9.0.
- You might want to convert a native BLAST-formatted scoring matrix to GCG-format.
Converting BLAST-Format scoring matrices to GCG format
GCG programs work with native
BLAST-formatted scoring matrices. Although converting BLAST-formatted scoring
matrices to GCG format is unnecessary, you may find it useful to do so. One
advantage GCG-formatted scoring matrices offer is that they allow you to set
specific gap creation and extension penalties within the scoring matrix file.
(If gap creation and extension penalties are not specified within a scoring
matrix file, programs determine default values on the fly.) In addition, GCG
by default assumes all native BLAST scoring matrices
are protein. Because gap creation and extension penalties are calculated
differently depending on if the matrix is nucleotide or protein, you may want
to convert the BLAST matrices to ensure they are the correct type.
To convert BLAST-Formatted scoring matrices to GCG format:
Type % reformat -COMParison scoring_matrix -NUCleotide or % reformat -COMParison scoring_matrix -PROtein.
TIP - Sometimes scoring matrices may be hard to edit because the lines wrap on your screen. To make your task easier, reformat the data file into columns using the command % reformat -COMParison -EQUALSformat scoring_matrix. Programs can read data files in this format as well as the regular format. (In the regular format, the sequences symbols are organized along the x axis (columns) and y axis (rows), where each symbol along the x axis is compared with each symbol along the y axis. The value of each pair of symbols compared is placed at the intersection of the appropriate row and column.)
Although it is not necessary, you can reformat a data file in columns back to its regular format using the command % reformat -COMParison scoring_matrix.
[Genhelp | Program Manual | User's Guide | Data Files | Databases | Release Notes ]
Technical
Support: support-us@accelrys.com, support-japan@accelrys.com,
or support-eu@accelrys.com
Copyright (c) 1982-2005
Accelrys Inc. All rights reserved.
Licenses and Trademarks:
Discovery Studio ®, SeqLab ®, SeqWeb ®, SeqMerge ®, GCG ®
and, the GCG logo are registered trademarks of Accelrys Inc.
All other product names
mentioned in this documentation may be trademarks, and if so, are trademarks or
registered trademarks of their respective holders and are used in this
documentation for identification purposes only.
